Step-Out RACE
Obtaining a full-length cDNA is a very important, and often very difficult step in gene discovery. Step-Out RACE technology allows fast isolation of cDNA ends, requires only first-strand cDNA synthesis, and is applicapable to total RNA (Matz et al., 1999a; Matz et al., 2003).
First-strand cDNA synthesis should be performed using any approach resulting in full-length-enriched cDNA flanked with known adapter sequences. Such cDNA can be prepared using well-known SMART approach (Matz et al., 1999; Zhu et al., 2001). Alternatively, suitable cDNA can be prepared using Mint method (Evrogen). First-strand cDNA is used for Step-Out PCR (Matz et al., 1999) with gene-specific primer (GSP) and specially designed Step-Out primer mix (SO-mix). The figure below details molecular events that occur during the Step-Out RACE procedure.
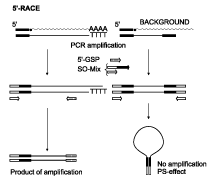 | Schematic outline of 5'-Step-Out RACE.Rectangles represent long inverted terminal repeats (ITRs) and correspondent primers. |
|---|
SO-mix is comprised of a long primer having 3'-half corresponding to the adapter sequence (specifically, to its distal half), and a 5'-portion that is incapable of annealing to the adapter. This 5'-portion is used for annealing of the second primer of the Step-Out mixture. In the PCR product, the non-matching 5'-portion of the new primer becomes added to the distal end of the original adapter, leading to a shift of the primer annealing site to the outside of the original amplicon, hence the name "Step-Out". Thus, inverted terminal repeats are generated on the background products that prevent background amplification owing to PCR suppression effect (Lukyanov et al., 1999).
The PCR suppression effect (PS-effect) relies on the fact that molecules flanked by inverted terminal repeats at least 40 bases long are amplified very inefficiently by single primer corresponding to the distal half of the repeat (Siebert et al., 1995; Chenchik et al., 1996), or by low concentrations of a long primer corresponding to the whole repeat sequence (Shagin et al., 1999). The PS-effect prohibits amplification of molecules lacking the annealing site for a primer of interest (Lukyanov et al., 1999). This is due to the equilibrium between productive primer annealing and non-productive self-annealing of the fragment's complementary ends, which arises during the annealing phase of each PCR cycle. In the above case, the equilibrium is markedly shifted towards self-annealing, providing the basis for the suppression effect.
Step-Out RACE assumes simultaneous RACE for 3'- and 5'-flanks, starting from a known fragment of cDNA sequence at least 20-30 bp long. The method is also applicable for 3'- and 5'-RACE starting from a partial protein sequence (at least 10 amino acids). In this case, degenerative primers are used for amplification of cDNA ends instead of gene-specific primers as used in the other cases (Matz et al., 1999b).
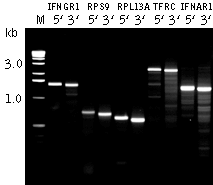 | Typical results of 5'- and 3'- Step-Out RACE.Placenta total RNA was used for the SMART first strand cDNA preparation and 5'- and 3'- RACE procedures with SO-mix and primers specific to IFNGR1( interferon gamma receptor 1); RPS9 (ribosomal protein S9); RPL13A (ribosomal protein L13a); TFRC (transferrin receptor); and IFNAR1 (interferon receptor 1) genes. |
|---|
The method produces target product using a single Step-Out PCR reaction, however non-specific amplification from the gene-specific primer often limits RACE power. An additional Step-Out PCR step using nested gene-specific primers may be required to resolve this problem.
A detailed protocol of the Step-Out RACE procedure can be found at (Matz et al., 2003).
References
- Chenchik A, Diachenko L, Moqadam F, Tarabykin V, Lukyanov S and Siebert PD (1996) Full-length cDNA cloning and determination of mRNA 5' and 3' ends by amplification of adaptor-ligated cDNA. Biotechniques 21, 526-534.
- Lukyanov KA, Gurskaya NG, Bogdanova EA and Lukyanov SA (1999) Selective suppression of polymerase chain reaction. Bioorganicheskaya Khimiya (Russ) 25, 163-170.
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. (1999a) Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Research 27(6): 1558-1560.
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. (1999b) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 17(10):969-73.
- Matz MV, Alieva NO, Chenchik A, Lukyanov S. (2003) Amplification of cDNA ends using PCR suppression effect and step-out PCR. Methods Mol Biol. 221: 41-49.
- Shagin DA, Lukyanov KA, Vagner LL and Matz MV (1999) Regulation of average length of complex PCR product. Nucleic Acids Res 27, e23.
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. (2001) Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30(4):892-897.
