Genome walking
Genome walking is a method to isolate flanking genomic segments (e.g. promoter regions) adjacent to a known sequence. We use the genome walking method based on the effect of PCR suppression by inverted terminal repeats (Lukyanov et al., 1994; Siebert et al., 1995; Lukyanov et al., 1999). Briefly, upon PCR initiation after the denaturation step, the single-stranded (ss) DNA fragments flanked by inverted terminal repeats (ITRs) may form either self-annealing "pan-like" structures (preventing the primer binding to its complementary binding sites and suppressing the PCR) or DNA/primer hybrid structures. In the latter case, if the primer corresponding to the outer part of ITR is used, the DNA synthesis by Taq-polymerase restores the original structure, ensuring the persistence of suppression during further PCR cycles.
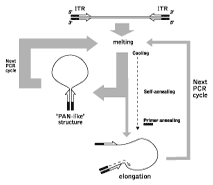 | Schematic outline of PCR suppression by inverted terminal repeats. |
|---|
The genome walking procedure is schematically outlined in the figure below. Uncloned genomic DNA is digested with various restriction endonucleases and ligated to long suppression adapters. The desired genomic region is amplified with a primer specific to the outer part of the suppression adapter and a gene-specific primer. Since the adapters are long and the adapter-specific primer is short (the sufficient ratio is 40 to 20 base pairs), the amplification of the whole pool from that single adapter-specific primer is effectively suppressed, and only the fragments of interest are generated during PCR. Obviously, both flanks of the known sequence – downstream as well as upstream – can be amplified: the direction depends exclusively upon the strand specificity of the gene-specific primer.
If a gene of interest contains multiple repeated elements or other complicated structures, we apply a specific approach that allows regulation of the length of PCR products and isolation of the longest fragment(s) of interest (Shagin et al., 1999).
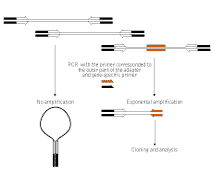 | Schematic outline of genome walking procedure. |
|---|
References
- Lukyanov SA, Gurskaya NG, Lukyanov KA, Tarabykin VS and Sverdlov ED (1994) Highly efficient subtractive hybridisation of cDNA. Bioorg. Khim., 20, 701-704 (Russ).
- Lukyanov KA, Gurskaya NG, Bogdanova EA and Lukyanov SA (1999) Selective suppression of polymerase chain reaction. Bioorganicheskaya Khimiya (Russ) 25, 163-170.
- Shagin DA, Lukyanov KA, Vagner LL and Matz MV (1999) Regulation of average length of complex PCR product. Nucleic Acids Res 27, e23.
- Siebert PD, Chenchik A, Kellog DE, Lukyanov KA and Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res., 23, 1087-1088.
