PCR amplification of long DNA fraction
In recent years, methods for PCR amplification of long DNA fragments have advanced significantly, especially with the invention of enzyme mixtures designed for "long and accurate PCR" (Barnes, 1994). Still, PCR tendency to amplify shorter fragments more efficiently than longer ones, most noticeably during the amplification of complex DNA mixtures containing more than one fragment, is well known. This tendency arises during the preparation of amplified full-length-enriched cDNA, during the amplification of cDNA ends, and during PCR-based genome walking. In all of these cases, the bias towards shorter DNA fragments may pose a significant problem during subsequent cloning and analysis of the PCR product. To shift the PCR preference towards longer products, we offer the PCR suppression-based method allowing amplification of long DNA fractions during PCR (Shagin et al., 1999).
The PS-effect occurs when the original ITR (inverted terminal repeat) is approximately double the length of the PCR primer. In this case, intramolecular annealing is much more efficient than primer annealing and amplification becomes completely (or for very long fragments, almost completely) suppressed (Lukyanov et al., 1994; Siebert et al., 1995).
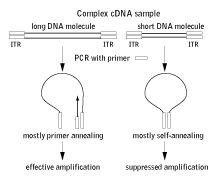 | Schematic outline of PCR suppression effect on long and short DNA molecules.
Rectangles represent long inverted terminal repeats (ITRs) and corresponding primers. |
|---|
When the ITRs equal the PCR primer in length, several factors (except the length of the fragment) influence the final outcome of the competition.
- GC-content of the ITR (and therefore of the primer): When the ITR and primer are represented by an oligo-dT stretch with a 5'-heel, only the shortest fragments (up to 300-400 bp) are suppressed, but with a very G/C-rich ITR and primer the suppression effect can extend to longer fragments as well.
- Primer concentration: Decrease of the primer concentration shifts the equilibrium towards intramolecular annealing. Therefore, the lower the primer concentration, the more pronounced the suppression. This parameter can be easily varied to regulate the degree of suppression.
Thus, by attaching ITRs to a complex DNA sample and amplifying it with a single ITR-specific primer at different concentrations, PCR preferences towards shorter products and longer products can be manipulated (Shagin et al., 1999). Using this approach it is possible to regulate the average length of the PCR product over a very wide range.
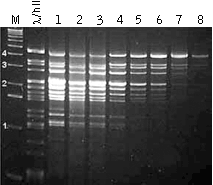 | Regulation of the average length of PCR product in the model system (λ DNA digested by HindII).Lane M – cDNA size markers; Lanes 1-8 – PCR product obtaining with various PCR parameters and primer length. |
|---|---|
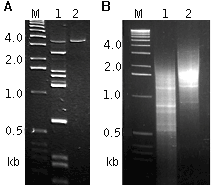 | Extraction of the longest fragments from a complex RACE product (A) and regulation of the average length of an amplified cDNA sample for library construction (B). |
References
- Lukyanov SA, Gurskaya NG, Lukyanov KA, Tarabykin VS and Sverdlov ED (1994) Highly efficient subtractive hybridisation of cDNA. Bioorg. Khim., 20, 701-704 (Russ).
- Shagin DA, Lukyanov KA, Vagner LL, Matz MV (1999) Regulation of average length of complex PCR product. Nucleic Acids Res., 27(18):e23.
- Siebert PD, Chenchik A, Kellog DE, Lukyanov KA and Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res., 23, 1087-1088.
