DSN-normalization of full-length-enriched cDNA
In an eukaryotic cell, the mRNA population constitutes approximately 1 of total RNA with the number of transcripts varying from several thousand to several tens of thousands. Normally, the high abundance transcripts (several thousand mRNA copies per cell) of as few as 5-10 genes account for 20% of the cellular mRNA. The intermediate abundance transcripts (several hundred copies per cell) of 500-2000 genes constitute about 40-60% of the cellular mRNA. The remaining 20-40 % of mRNA is represented by rare transcripts (from one to several dozen mRNA copies per cell) (Alberts et al., 1994). Such an enormous difference in abundance complicates large-scale transcriptome analysis, which results in recurrent sequencing of more abundant cDNAs.
cDNA normalization decreases the prevalence of high abundance transcripts and equalizes transcript concentrations in a cDNA sample, thereby dramatically increasing the efficiency of sequencing and rare gene discovery.
Normalization is utilized to enhance the gene discovery rate of a cDNA library and facilitate the identification and analysis of rare transcripts. This approach is imperative for transcriptom sequencing, and useful in other applications, such as functional screening, construction of specific RNA libraries, and Transcript End Sequence Profiling.
cDNA normalization using duplex-specific nuclease (DSN) is a highly efficient approach that can be applied for normalization of full-length-enriched cDNA (Zhulidov et al., 2004; Zhulidov et al., 2005). The resulting cDNA contains equalized abundance of different transcripts and can be used for construction of cDNA libraries and for direct sequencing, including high-throughput sequencing on the next generation sequencing platforms (Roche/454, ABI/SOLiD or Illumina/Solexa).
Method overview
cDNA normalization using duplex-specific nuclease (DSN) is a highly efficient approach that can be applied for normalization of full-length-enriched cDNA (Zhulidov et al., 2004; Zhulidov et al., 2005). The method is based on nucleic acid hybridization kinetics (Young and Anderson, 1985) and unique properties of the duplex-specific nuclease (DSN) specific to the double-stranded (ds) DNA (Shagin et al., 2002).
DSN-normalization is performed prior to library cloning. After denaturation of ds cDNA flanked with known adapters, it is subjected to renaturation. During renaturation, abundant transcripts convert to the ds form more effectively than those that are less frequent. Thus, two fractions are formed, specifically, a ds-fraction of abundant cDNA and a normalized single-stranded (ss) cDNA. The ds cDNA fraction is then degraded by DSN.
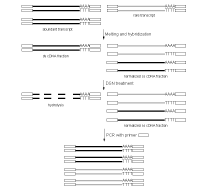 | Schematic outline of DSN-normalization.Black lines represent abundant transcripts, grey lines – rare transcripts. Rectangle represents adapter sequence and its complement. |
|---|
DSN is an enzyme from Red King (Kamchatka) crab that displays a strong preference for cleaving ds DNA compared to ss-DNA and RNA, irrespective of the sequence length. DSN is stable under elevated temperatures (Shagin et al., 2002). Maximal DSN activity is observed at 60-65°C, and about 25% of the activity is retained after incubation at 70°C for 20 min. Owing to DSN thermostability, ds DNA degradation is performed under conditions of cDNA renaturation that prevent the formation of secondary structures and non-specific hybridization involving adapter sequences within the ss cDNA fraction.
The remaining normalized ss DNA is amplified by PCR. PCR primers and conditions are optimized to minimize the PCR tendency to amplify shorter fragments more efficiently than longer ones. Normalized cDNA can then be used for library cloning or sequencing.
cDNA suitable for normalization can be prepared on the basis of total or poly(A)+ RNA and should contain known adapter sequences at both ends for PCR amplification. The quality of the RNA is crucial, especially when construction of full-length enriched cDNA library is a goal. The flanking sequences can be introduced to the cDNA ends during cDNA synthesis by various means, for example, adapter ligation or during cDNA synthesis using the template-switching approach.
DSN-normalization has been successfully applied to various animal and plant models (see Bogdanova et al., 2008 for review). The flexibility of this normalization procedure allows simple modifications for various purposes. Detailed protocols of DSN-normalization modifications are available in the book Nucleic Acids Hybridization Modern Applications (Shcheglov et al., 2007), and Current Protocols (Bogdanova et al., 2010).
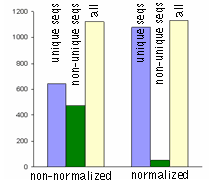 | Normalization significantly increases gene discovery rate of cDNA library.Transcript distribution in standard and normalized cDNA libraries from Aplysia neurons: blue – unique sequences; green – non-unique sequences, yellow – all sequences. |
|---|
 |  |  | |
|---|---|---|---|
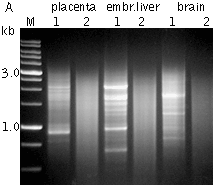
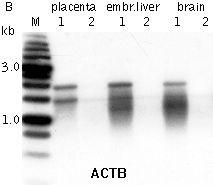
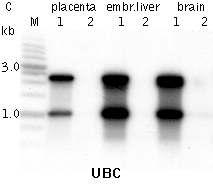
Typical cDNA normalization result.(A) Agarose/EtBr gel-electrophoresis of non-normalized (1) and normalized (2) human cDNA samples; (B, C) concentration of abundant transcripts in these samples revealed by Virtual Northern blot. ACTB – β-actin; UBC – ubiquitin C; M, 1-kb DNA size markers (SibEnzyme); embr. – embryonic. | |||
References:
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 3rd ed., Garland Publishing, New York. 1994; .
- Bogdanova EA, Shagin DA, Lukyanov SA. Normalization of full-length enriched cDNA. Mol Biosyst. 2008; 4 (3):205-12. / pmid: 18437263
- Bogdanova EA, Shagina I, Barsova EV, Kelmanson I, Shagin DA, Lukyanov SA. Normalizing cDNA libraries. Curr Protoc Mol Biol. 2010; Chapter 5 :Unit 5.12.1-27. / pmid: 20373503
- Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002; 12 (12):1935-42. / pmid: 12466298
- Shcheglov A, Zhulidov P, Bogdanova E, Shagin D. Normalization of cDNA Libraries. In: Nucleic Acids Hybridization: Modern Applications. Buzdin, Anton; Lukyanov, Sergey (Eds.) ISBN: 978-1-4020-6039-7. 2007; 97-124.
- Young BD, Anderson M. Quantitative analysis of solution hybridisation. In Nucleic Acids Hybridisation, a practical approach (eds. Hames, B.D. and Higgins, S.J.) IRL Press, Oxford-Washington DC. 1985; 47-71.
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Shagina IA, Wagner LL, Khazpekov GL, Kozhemyako VV, Lukyanov SA, Shagin DA. A method for the preparation of normalized cDNA libraries enriched with full-length sequences. Bioorg Khim. 2005; 31 (2):186-94. / pmid: 15889793
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, Shagin DA. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004; 32 (3):e37. / pmid: 14973331
